Context and prior work
Prior work includes Darwin from Xie et al. (Xie et al. 2023), that finetuned the Llama-7B model (Touvron et al. 2023) on several chemistry tasks. Other works include ChemLLM (Zhang et al. 2024) from Zhang et al. that finetuned the InternLM2-Base-7B model (Cai et al. 2024) on 9 different tasks. Other works followed a similar line, with truly generalist chemical LLMs still missing from the literature.
Why this paper matters?
This is one of the few attempts to train a chemistry-specific foundation models from the pre-training stage, all the way to fine-tuning. The authors showed that specialized data, can help relatively small models like Llama-13B outperform frontier models like GPT-4 on specialized tasks. While again, the tasks are extremely specific, the sheer scale of 34B tokens in the pretraining is important, since it is unprecedented.
Model performance
The model seems to exhibit good zero-shot performance compared to GPT-4 on the molecule captioning tasks. The authors indicated a similar performance increase for other tasks like SMILES-to-IUPAC, which is at 0% for GPT-4, and at 4% for ChemDFM. While in the real world, and when compared to more specialist models like STOUT, the model does not provide significant utility.
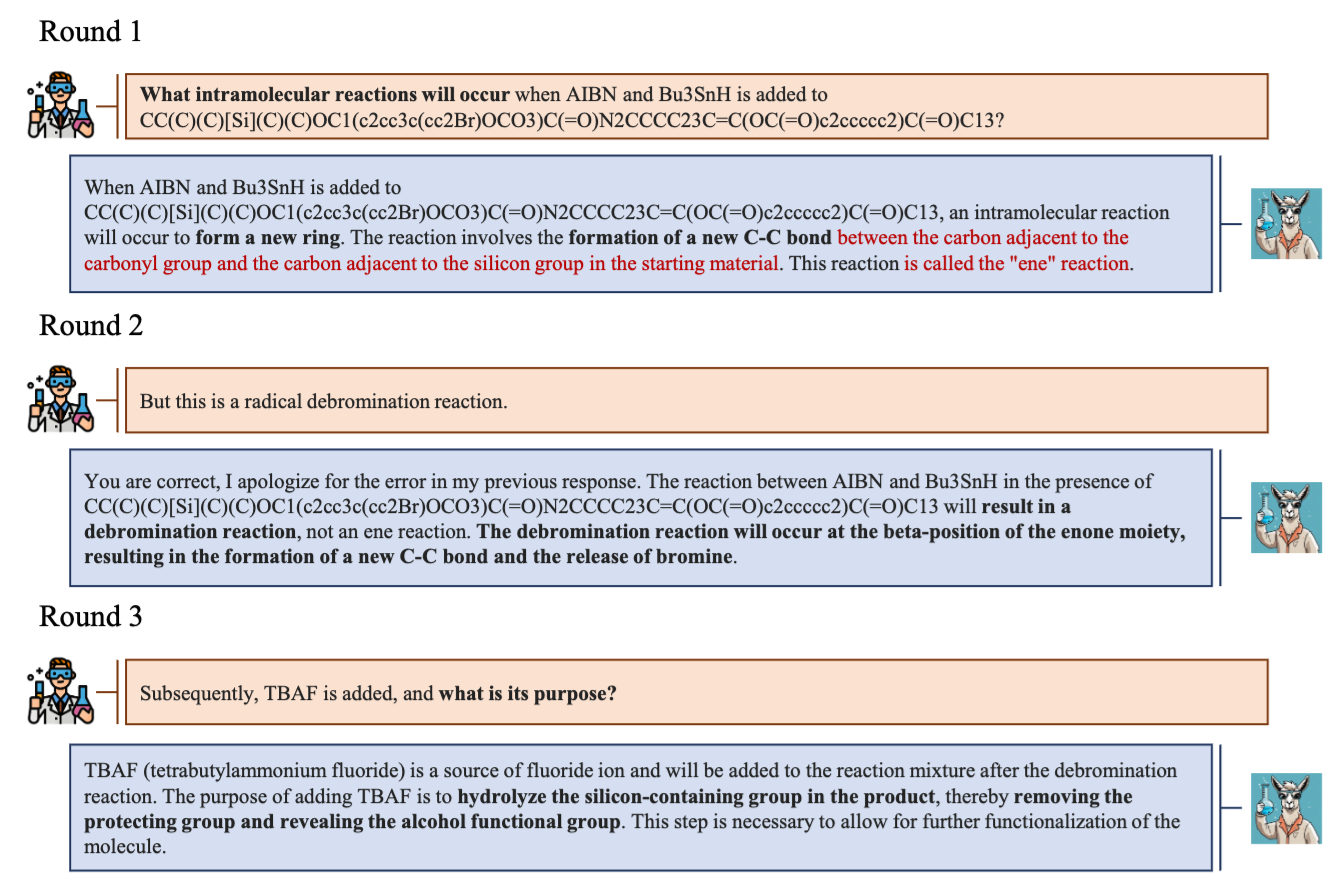
Moreover, they showed that the model exhibits good conversational capabilities, and the ability to correct its mistakes. For instance, in the snipped below, the model improves its wrong answer upon a correction coming from the user.
Limitations
The authors were among the first to perform full pretraining on a sizable corpus, but the shortcomings of this work lie in the lack of ablations. For example, while a performance exceeding the one of GPT-4 is promising for the open-source community in general, it is perhaps a very narrow set of tasks. GPT-4 outperformed ChemDFM on the 10-shot task, indicating that just with a simple prompting technique as in-context learning, the frontier model outperforms ChemDFM on the majority of metrics.
Moreover, ablations of the pre-training should be performed. It is not fully understood from this paper whether the pretraining helped or not, or just fine-tuning is responsible for the spurt in the zero-shot performance.